Note: This article first appeared in Research Europe and Research Professional News.
The Covid-19 pandemic has triggered an explosion of knowledge, with more than 200,000 papers published to date. At one point last year, scientific output on the topic was doubling every 20 days. This huge growth poses big challenges for researchers, many of whom have pivoted to coronavirus research without experience or preparation.
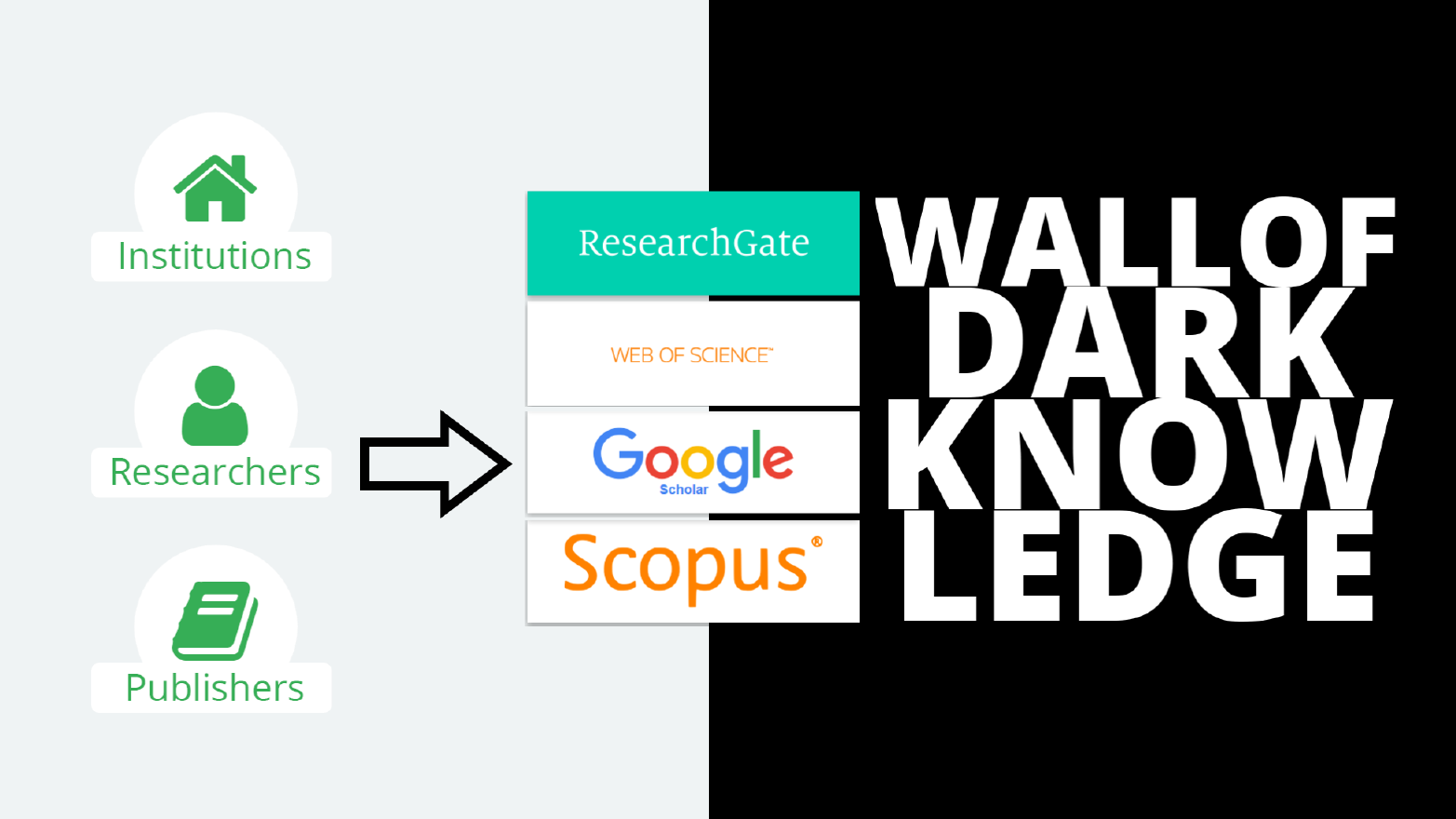
Mainstream academic search engines are not built for such a situation. Tools such as Google Scholar, Scopus and Web of Science provide long, unstructured lists of results with little context.
These work well if you know what you are looking for. But for anyone diving into an unknown field, it can take weeks, even months, to identify the most important topics, publication venues and authors. This is far too long in a public health emergency.
The result has been delays, duplicated work, and problems with identifying reliable findings. This lack of tools to provide a quick overview of research results and evaluate them correctly has created a crisis in discoverability itself.
The pandemic has highlighted this, but with three million research papers published each year, and a growing diversity of other outputs such as datasets, discoverability has become a challenge in all disciplines.
For years a few large companies have dominated the market for discovery systems: Google Scholar; Microsoft’s soon-to-be-closed Academic; Clarivate’s Web of Science, formerly owned by Thomson Reuters; and Elsevier’s Scopus.
But investment hasn’t kept pace with the growth of scientific knowledge. What were once groundbreaking search engines have only been modestly updated, so that mainstream discovery systems are now of limited use.
This would not be a problem if others could build on companies’ search indices and databases. But usually they can’t.
A new openness
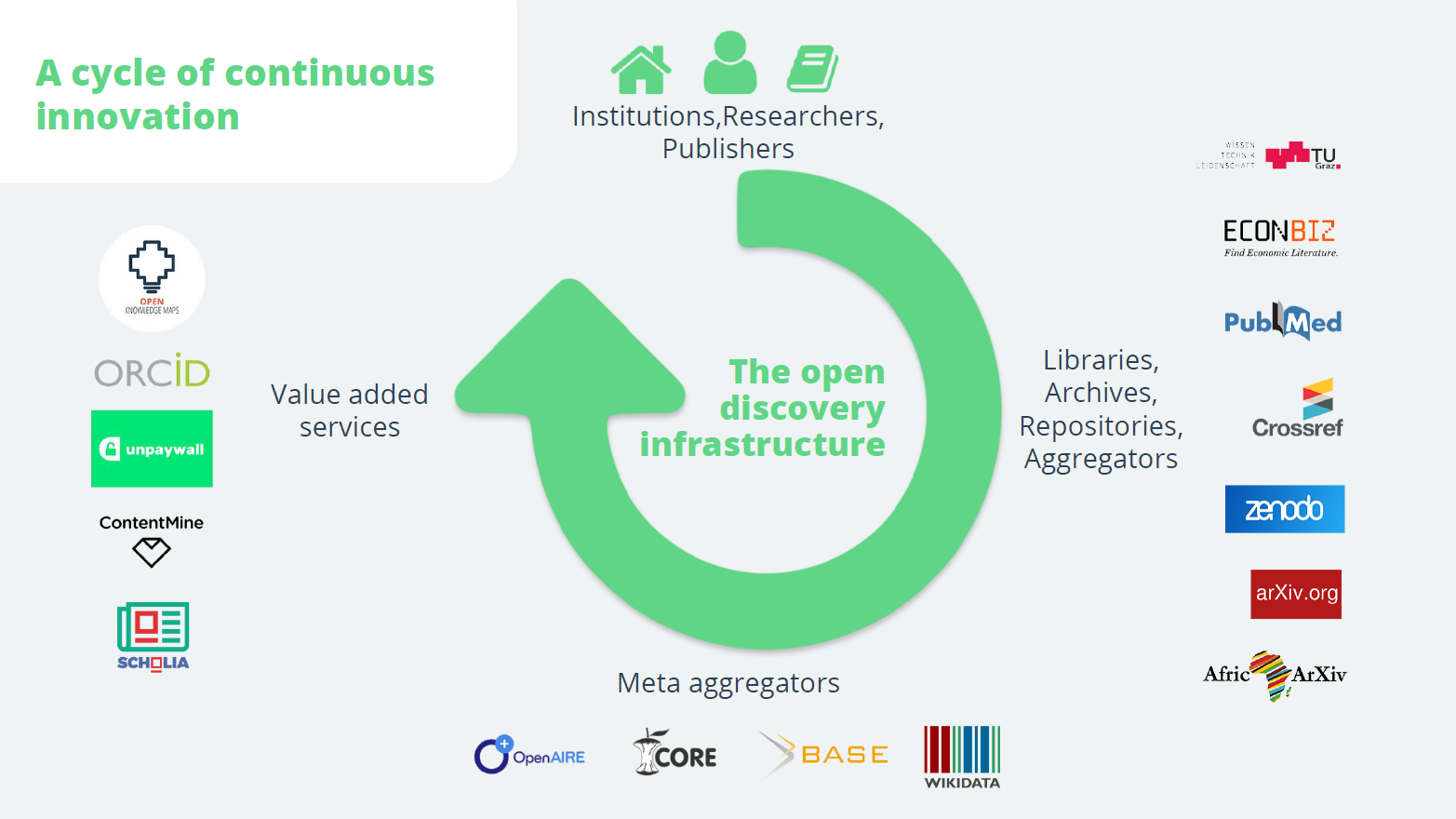
In the shadows of these giants, however, an alternative discovery infrastructure has been created, built on thousands of public and private archives, repositories and aggregators, and championed by libraries, non-profit organisations and open-source software developers. Unlike the commercial players, these systems make their publication data and metadata openly available.
Building on these, meta-aggregators such as Base, Core and OpenAIRE have begun to rival and in some cases outperform the proprietary search engines. Their openness supports a rich ecosystem of value-added services, such as the visual discovery system Open Knowledge Maps, which I founded, or the open-access service Unpaywall.
This open infrastructure has become the strongest driver of innovation in discovery, enabling the quick development of a variety of discovery tools during the pandemic. Technologies such as semantic search, recommendation systems and text and data mining are increasingly available.
Many open systems, though, are not sustainably funded. Some of the most heavily used make ends meet with a tiny core team. Half, including Open Knowledge Maps, rely on volunteers to provide basic services.
The funding options for non-profit organisations and open-source projects are very limited. Most rely on research grants, which are meant as a jumping off point, not a long-term solution.
The academic community needs to step up and secure the future of this crucial infrastructure. The shape of research infrastructure depends on institutions’ buying decisions. If most of the money goes to closed systems, these will prevail.
A first step would be to create dedicated budget lines for open infrastructures. The initial investment would be relatively small, as their membership fees are usually orders of magnitude cheaper than the license fees of their proprietary counterparts. Over time, strengthening open infrastructure will enable research institutions to cancel their proprietary products.
It’s not just about money. Open infrastructures do not lock institutions into closed systems, and save them from selling off their researchers’ user data, an issue gaining prominence as large commercial publishers become data analytics businesses.
The coronavirus pandemic has shown that the challenges of our globalised world demand international collaboration. That requires building on each others’ knowledge.
This is not possible with closed and proprietary discovery infrastructures that have fallen behind the growth of scientific knowledge. Instead, we need to guarantee the sustainability of the open discovery infrastructure, so that we can rely on it for today’s and tomorrow’s challenges.




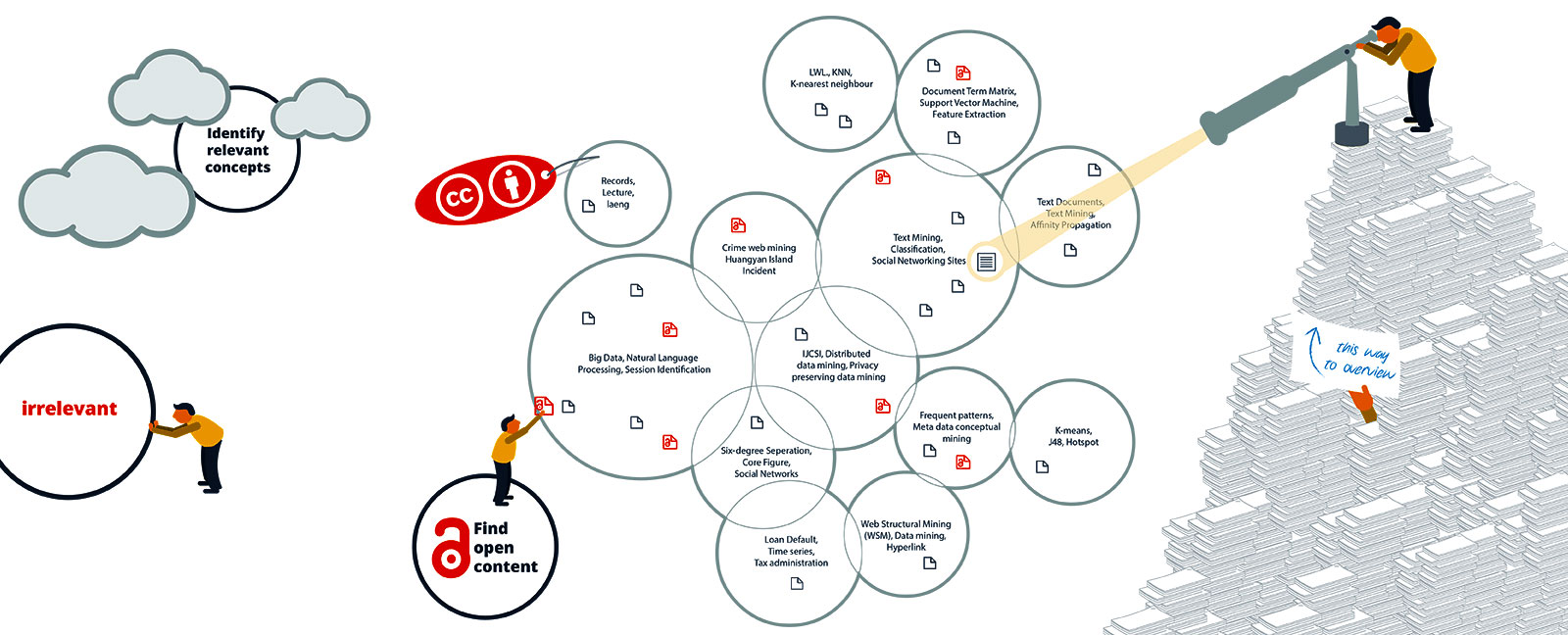
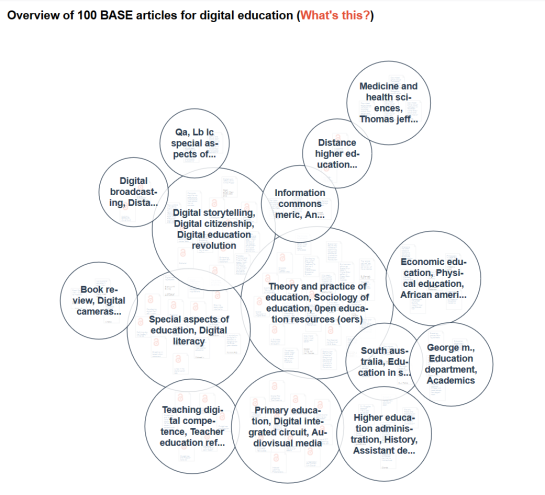
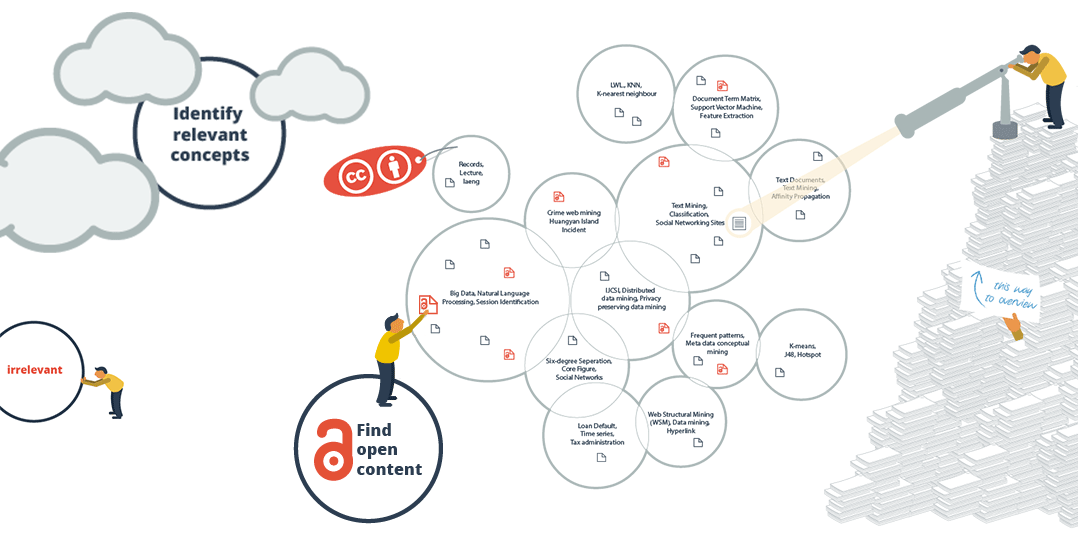
 The sub-areas also make you aware of the terminology in a field. This information alone may take weeks to find out. How much time have you already lost to searching without knowing the best search terms? In addition, the knowledge map enables users to separate the wheat from the chaff with respect to their current information need. For an ambiguous search term for example, the different meanings are sorted into separate areas.
The sub-areas also make you aware of the terminology in a field. This information alone may take weeks to find out. How much time have you already lost to searching without knowing the best search terms? In addition, the knowledge map enables users to separate the wheat from the chaff with respect to their current information need. For an ambiguous search term for example, the different meanings are sorted into separate areas.